
Connecting Solids in Pipelines#
Our pipelines wouldn't be very interesting if they were limited to solids acting in isolation from each other. Pipelines are useful because they let us connect solids into arbitrary DAGs of computation.
Dagster pipelines model a dataflow graph. In data pipelines, the reason that a later step comes after an earlier step is almost always that it uses data produced by the earlier step. Dagster models these dataflow dependencies with inputs and outputs.
Let's Get Serial#
We'll expand the pipeline we worked with in the first section of the tutorial into two solids:
- The first solid will read the cereal dataset from the CSV and return it as an output.
- The second solid will consume the cereals produced by the first solid and find the most caloric cereal.
import csv
import os
from dagster import execute_pipeline, pipeline, solid
@solid
def load_cereals(context):
csv_path = os.path.join(os.path.dirname(__file__), "cereal.csv")
with open(csv_path, "r") as fd:
cereals = [row for row in csv.DictReader(fd)]
context.log.info(f"Found {len(cereals)} cereals".format())
return cereals
@solid
def sort_by_calories(context, cereals):
sorted_cereals = list(
sorted(cereals, key=lambda cereal: cereal["calories"])
)
context.log.info(f'Most caloric cereal: {sorted_cereals[-1]["name"]}')
@pipeline
def serial_pipeline():
sort_by_calories(load_cereals())
You'll see that we've modified our existing load_cereals solid to return an output, in this case
the list of dicts representing the cereals dataset.
We've defined our new solid, sort_by_calories, to take a user-defined input, cereals, in
addition to the system-provided context object.
We can use inputs and outputs to connect solids to each other. Here we tell Dagster that:
load_cerealsdoesn't depend on the output of any other solid.sort_by_caloriesdepends on the output ofload_cereals.
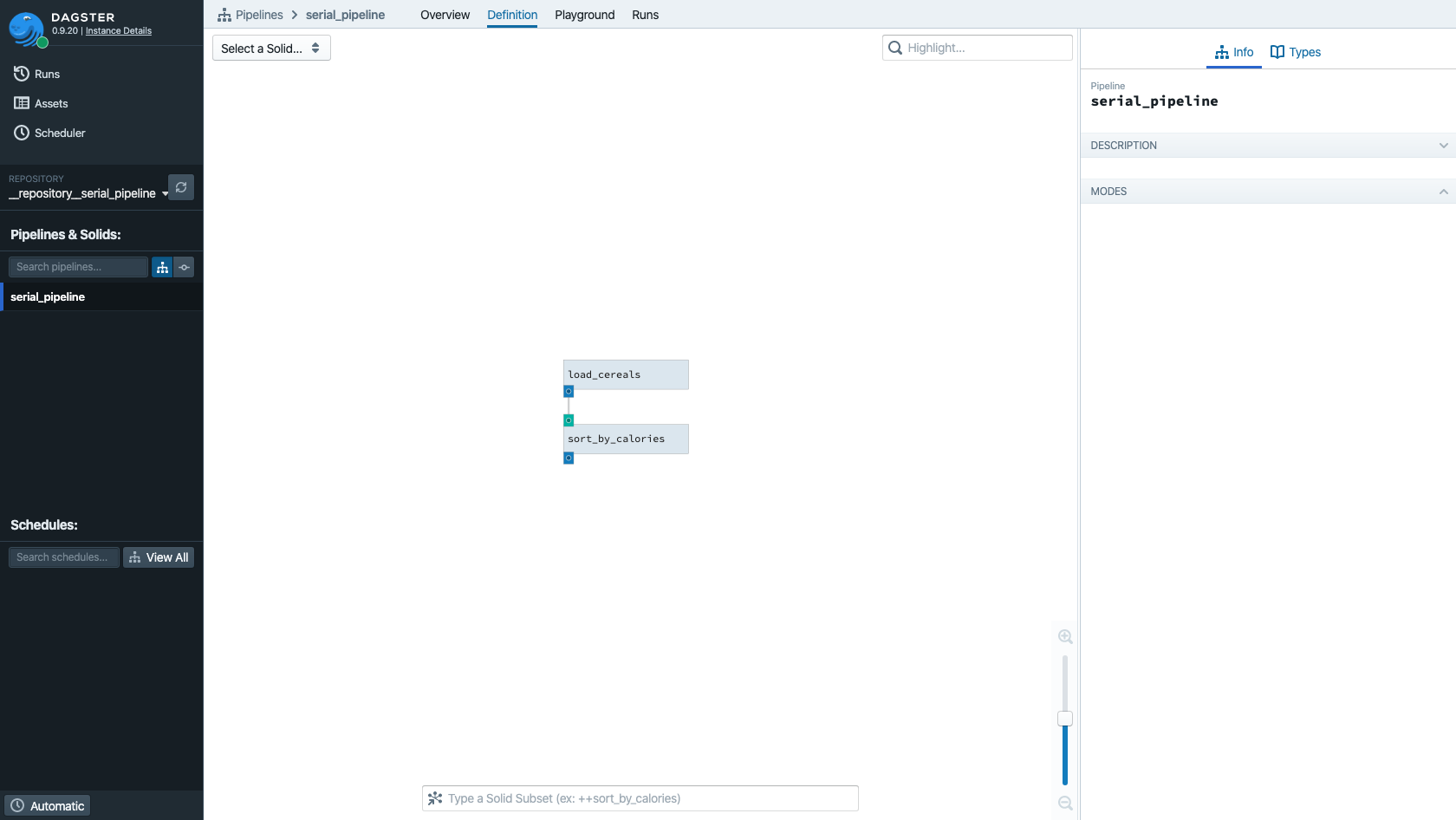
Let's visualize this pipeline in Dagit:
dagit -f serial_pipeline.py
Navigate to http://127.0.0.1:3000/pipeline/serial_pipeline/ or choose "serial_pipeline" from the left sidebar:
A More Complex DAG#
Solids don't need to be wired together serially. The output of one solid can be consumed by any number of other solids, and the outputs of several different solids can be consumed by a single solid.
@solid
def load_cereals(_):
dataset_path = os.path.join(os.path.dirname(__file__), "cereal.csv")
with open(dataset_path, "r") as fd:
cereals = [row for row in csv.DictReader(fd)]
return cereals
@solid
def sort_by_calories(_, cereals):
sorted_cereals = list(
sorted(cereals, key=lambda cereal: cereal["calories"])
)
most_calories = sorted_cereals[-1]["name"]
return most_calories
@solid
def sort_by_protein(_, cereals):
sorted_cereals = list(
sorted(cereals, key=lambda cereal: cereal["protein"])
)
most_protein = sorted_cereals[-1]["name"]
return most_protein
@solid
def display_results(context, most_calories, most_protein):
context.log.info(f"Most caloric cereal: {most_calories}")
context.log.info(f"Most protein-rich cereal: {most_protein}")
@pipeline
def complex_pipeline():
cereals = load_cereals()
display_results(
most_calories=sort_by_calories(cereals),
most_protein=sort_by_protein(cereals),
)
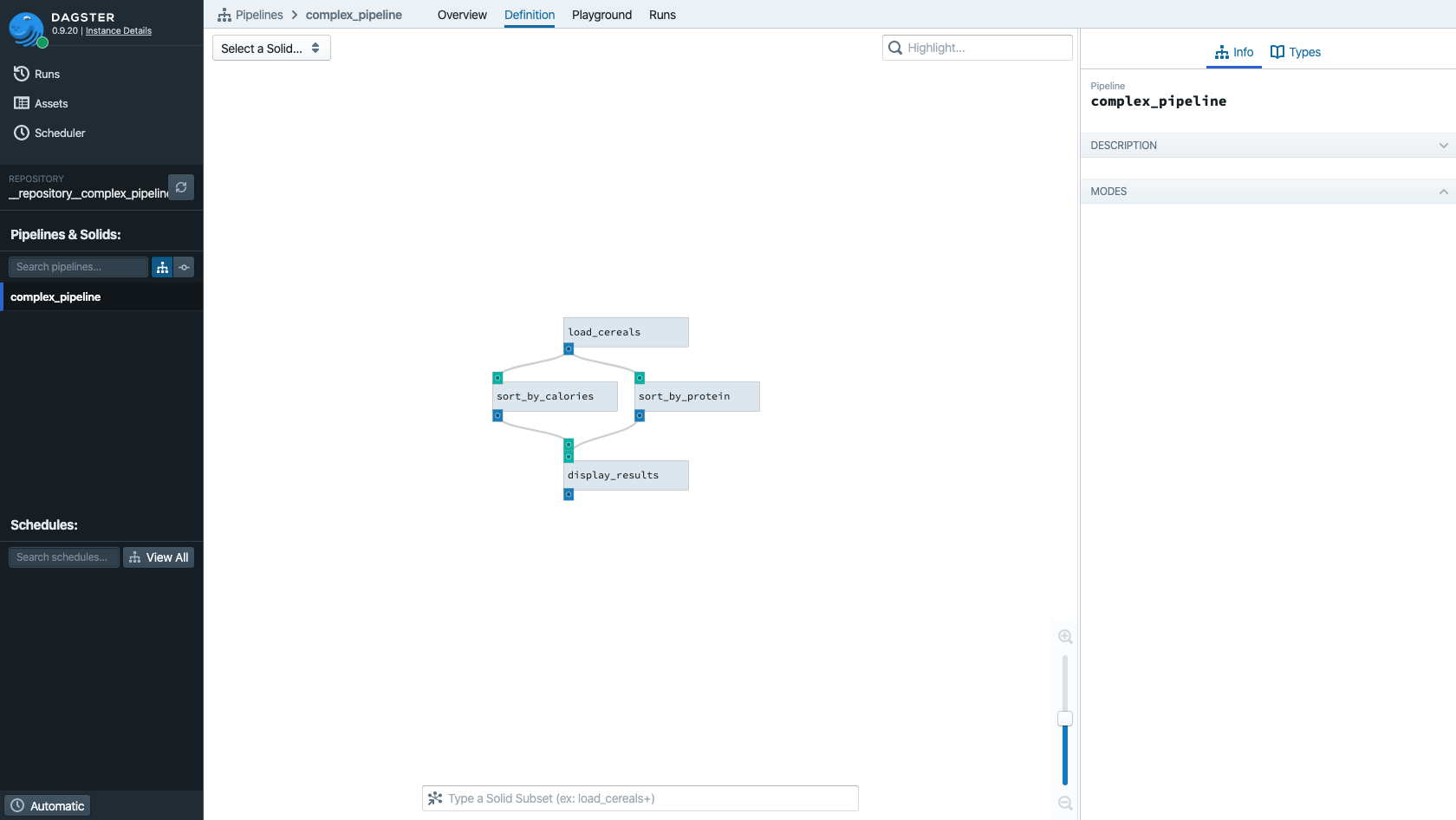
First we introduce the intermediate variable cereals into our pipeline definition to represent the
output of the load_cereals solid. Then we make both sort_by_calories and sort_by_protein
consume this output. Their outputs are in turn both consumed by display_results.
Let's visualize this pipeline in Dagit:
dagit -f complex_pipeline.py
When you execute this example from Dagit, you'll see that load_cereals executes first, followed by
sort_by_calories and sort_by_protein—in any order—and that display_results
executes last, only after sort_by_calories and sort_by_protein have both executed.
In more sophisticated execution environments, sort_by_calories and sort_by_protein could execute
not just in any order, but at the same time, since they don't depend on each other's
outputs—but both would still have to execute after load_cereals (because they depend on its
output) and before display_results (because display_results depends on both of their outputs).
We'll write a simple test for this pipeline showing how we can assert that all four of its solids executed successfully.
def test_complex_pipeline():
res = execute_pipeline(complex_pipeline)
assert res.success
assert len(res.solid_result_list) == 4
for solid_res in res.solid_result_list:
assert solid_res.success